A crew of researchers from Google Analysis and UC Santa Cruz launched DeepSomatic, an AI mannequin that identifies most cancers cell genetic variants. In analysis with Youngsters’s Mercy, it discovered 10 variants in pediatric leukemia cells missed by different instruments. DeepSomatic has a somatic small variant caller for most cancers genomes that works throughout Illumina brief reads, PacBio HiFi lengthy reads, and Oxford Nanopore lengthy reads. The tactic extends DeepVariant, detects single nucleotide variants and small insertions and deletions in complete genome and complete exome information, and helps tumor regular and tumor solely workflows, together with FFPE fashions.
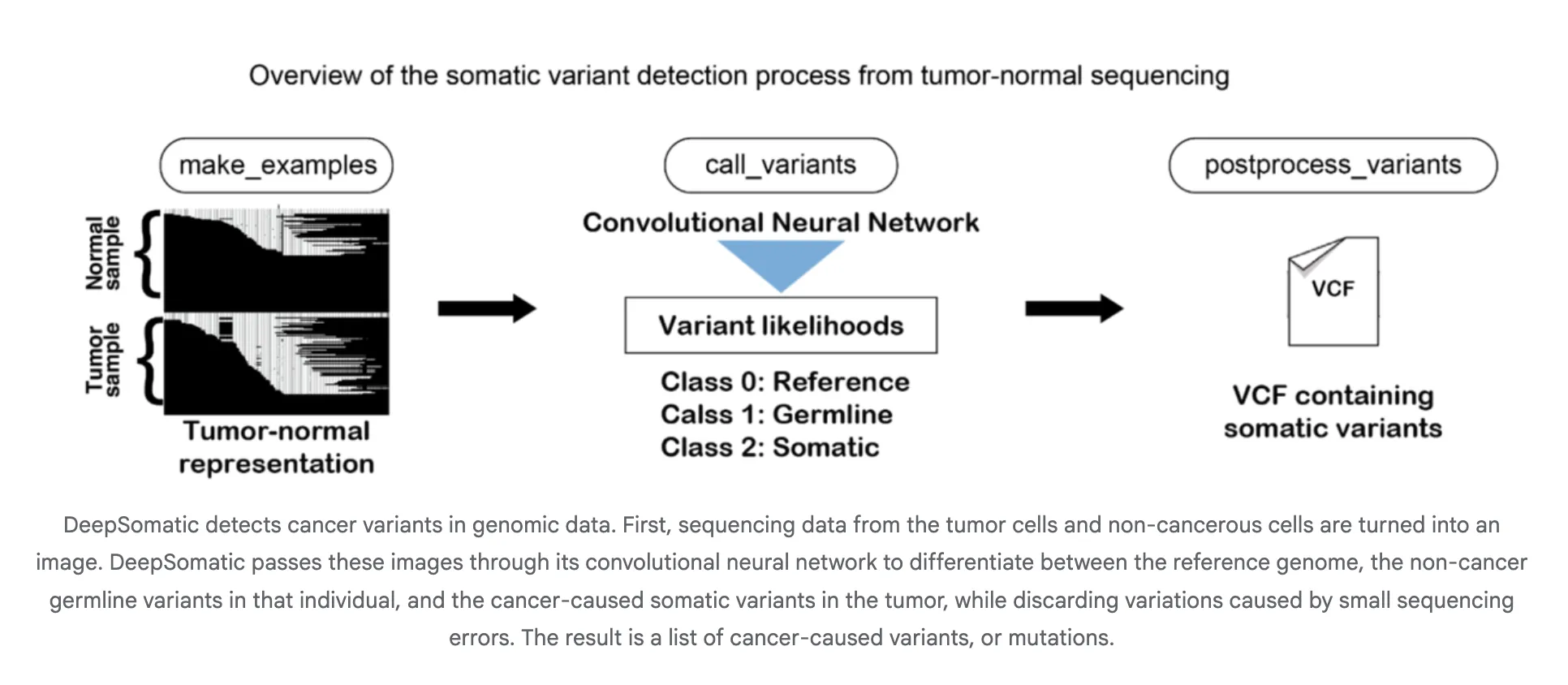

How It Works?
DeepSomatic converts aligned reads into picture like tensors that encode pileups, base qualities, and alignment context. A convolutional neural community classifies candidate websites as somatic or not and the pipeline emits VCF or gVCF. This design is platform agnostic as a result of the tensor summarizes native haplotype and error patterns throughout applied sciences. Google researchers describe the method and its deal with distinguishing inherited and bought variants together with tough samples similar to glioblastoma and pediatric leukemia.
Datasets and Benchmarking
Coaching and analysis use CASTLE, Most cancers Requirements Lengthy learn Analysis. CASTLE accommodates 6 matched tumor and regular cell line pairs that have been complete genome sequenced on Illumina, PacBio HiFi, and Oxford Nanopore. The analysis crew releases benchmark units and accessions for reuse. This fills a spot in multi know-how somatic coaching and testing assets.
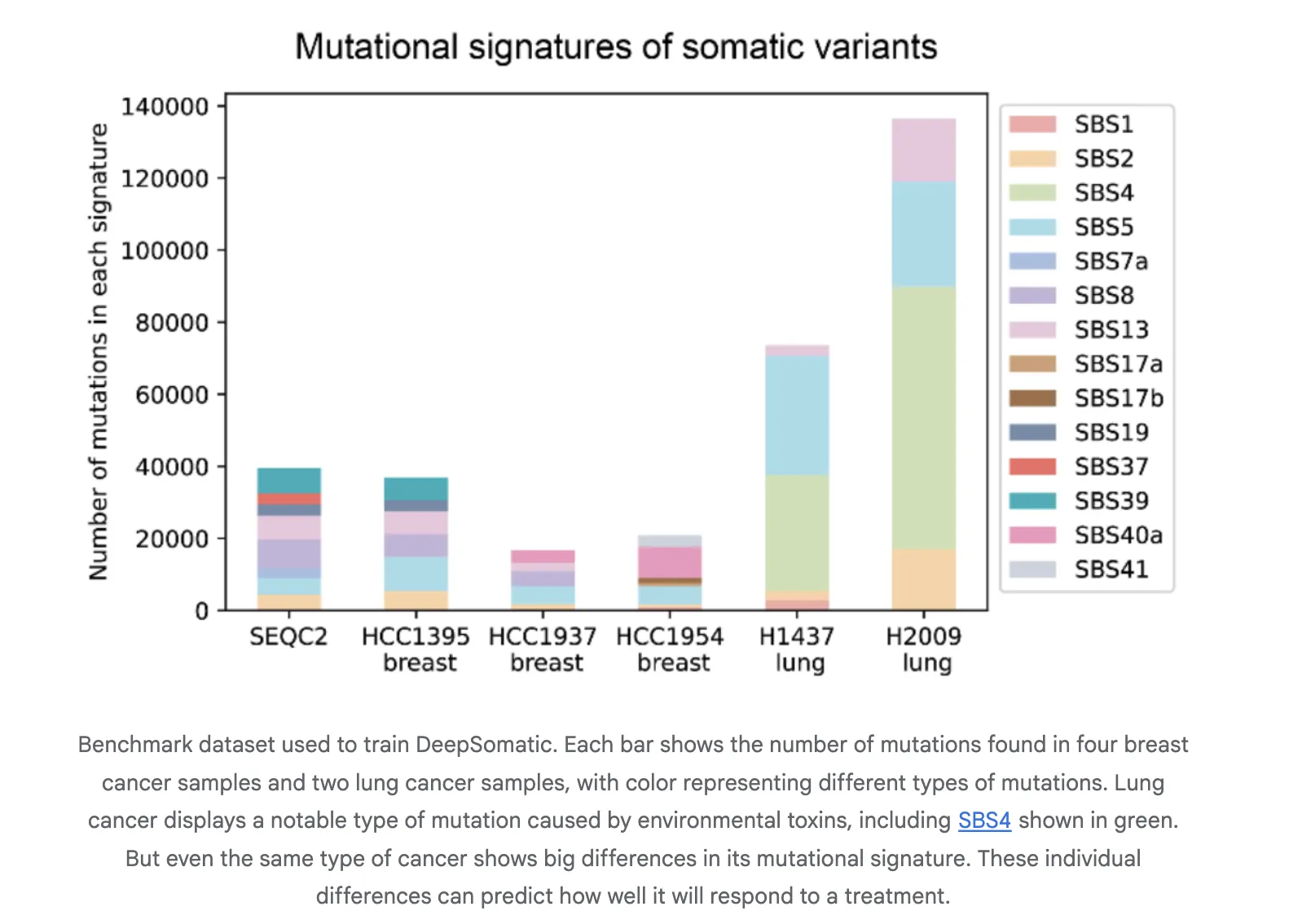

Reported Outcomes
The analysis crew report constant features over broadly used strategies in each single nucleotide variants and indels. On Illumina indels, the following greatest technique is about 80 p.c F1, DeepSomatic is about 90 p.c. On PacBio indels, the following greatest technique is underneath 50 p.c, DeepSomatic is above 80 p.c. Baselines embody SomaticSniper, MuTect2, and Strelka2 for brief reads and ClairS for lengthy reads. The examine stories 329,011 somatic variants throughout the reference traces and a further preserved pattern. Google analysis crew stories that DeepSomatic outperforms present strategies with explicit energy on indels.
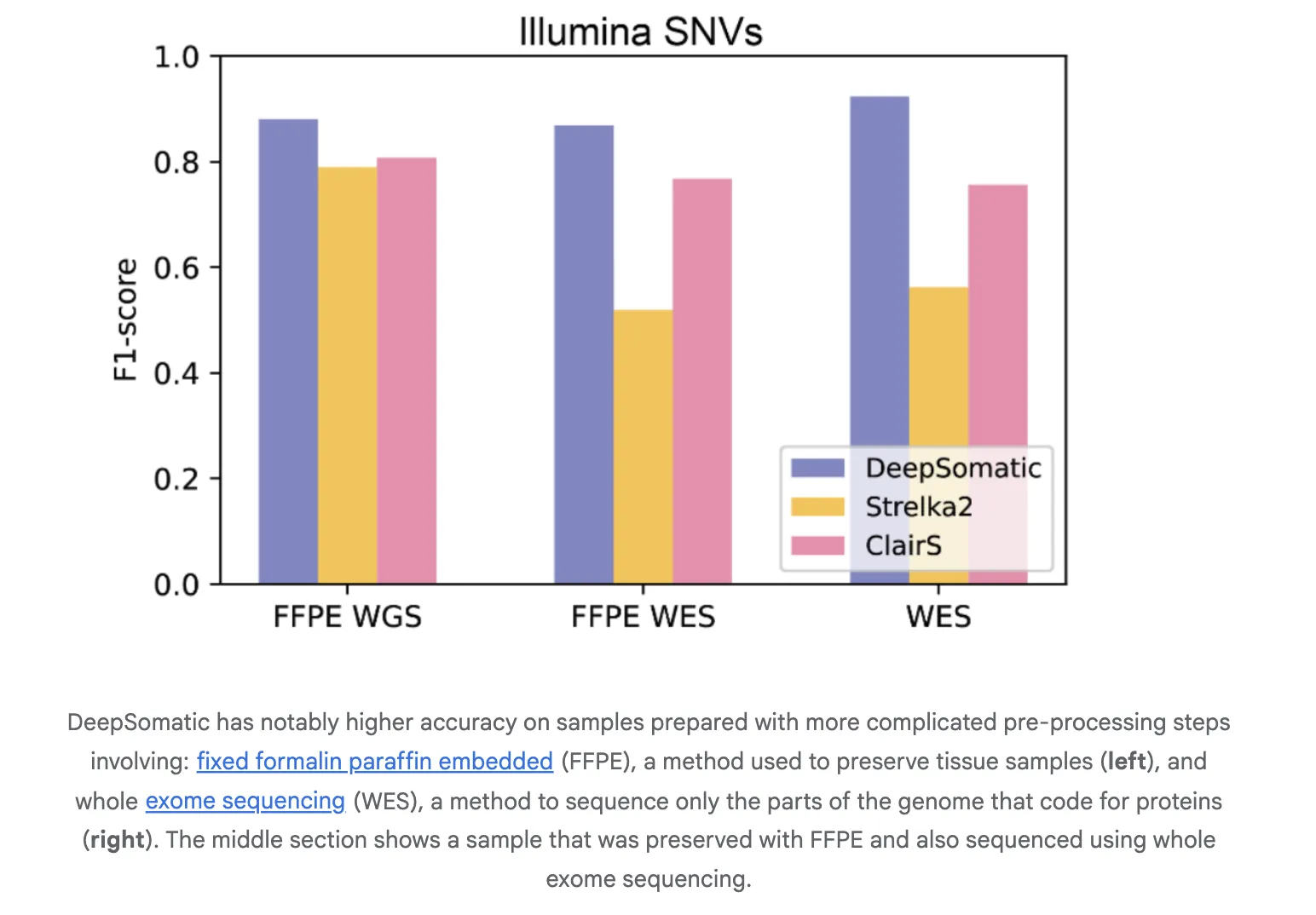

Generalization to Actual Samples
The analysis crew evaluates switch to cancers past the coaching set. A glioblastoma pattern reveals restoration of identified drivers. Pediatric leukemia samples take a look at the tumor solely mode the place a clear regular isn’t obtainable. The instrument recovers identified calls and stories extra variants in that cohort. These research point out the illustration and coaching scheme generalize to new illness contexts and to settings with out matched normals.
Key Takeaways
- DeepSomatic detects somatic SNVs (single nucleotide variants) and indels throughout Illumina, PacBio HiFi, and Oxford Nanopore, and builds on the DeepVariant methodology.
- The pipeline helps tumor regular and tumor solely workflows, consists of FFPE WGS and WES fashions, and is launched on GitHub.
- It encodes learn pileups as picture like tensors and makes use of a convolutional neural community to categorise somatic websites and emit VCF or gVCF.
- Coaching and analysis use the CASTLE dataset with 6 matched tumor regular cell line pairs sequenced on three platforms, with benchmarks and accessions supplied.
- Reported outcomes present about 90 p.c indel F1 on Illumina and above 80 p.c on PacBio, outperforming frequent baselines, with 329,011 somatic variants recognized throughout reference samples.
DeepSomatic is a practical step for somatic variant calling throughout sequencing platforms, the mannequin retains DeepVariant’s picture tensor illustration and a convolutional neural community, so the identical structure scales from Illumina to PacBio HiFi to Oxford Nanopore with constant preprocessing and outputs. The CASTLE dataset is the fitting transfer, it provides matched tumor and regular cell traces throughout 3 applied sciences, which strengthens coaching and benchmarking and aids reproducibility. Reported outcomes emphasize indel accuracy, about 90% F1 on Illumina and greater than 80% on PacBio in opposition to decrease baselines, which addresses an extended working weak spot in indel detection. The pipeline helps WGS and WES, tumor regular and tumor solely, and FFPE, which matches actual laboratory constraints.
Take a look at the Technical Paper, Technical particulars, Dataset and GitHub Repo. Be at liberty to take a look at our GitHub Web page for Tutorials, Codes and Notebooks. Additionally, be at liberty to comply with us on Twitter and don’t neglect to hitch our 100k+ ML SubReddit and Subscribe to our E-newsletter. Wait! are you on telegram? now you possibly can be part of us on telegram as effectively.


